hicValidateLocations¶
hicValidateLoops is a tool to compare the detect loops from hicDetectLoops (or from any other software as long as the data format is followed, see below) with known peak protein locations to validate if the computed loops do have the expected anchor points. Loops are usually bound by CTCF or Cohesin, therefore it is important to know if the detect loops have protein peaks at their X and Y position.
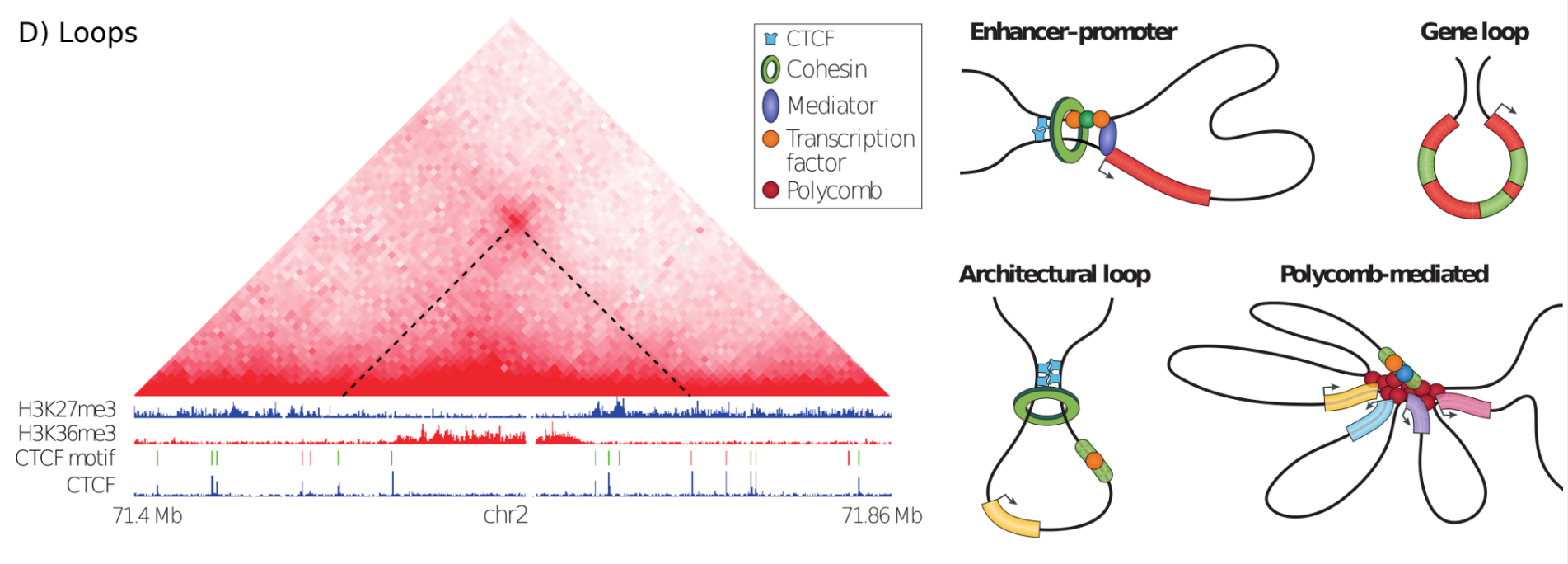
Loops in Hi-C, graphic from Bonev & Cavalli, Nature Reviews Genetics 2016¶
Data format¶
The data format of hicDetectLoops output is:
chr_x start_x end_x chr_y start_y end_y p-value
As protein input narrowPeak or broadPeak files are tested. However, as long as the protein data contains in the first three columns the chromosome, start and end it should work too.